Neuron Arrays and Weight Matrices
Simbrain originated as a way to focus intuition on loose or “free”-standing neurons and their connections, which model neuroscience or else show how the connection to neuroscence works in an intuitive way. However most neural networks software use vectors and matrices to represent neural networks, which make computations far more efficient.
In Simbrain 4 array-based approaches have been incorporated (using Smile matrix objects), and are integrated in such a way that they can be used alongside all the existing machinery, using similar GUI visualizations. Arrays and matrices provide efficient operations for large-scale neural network simulations and support modern machine learning workflows.
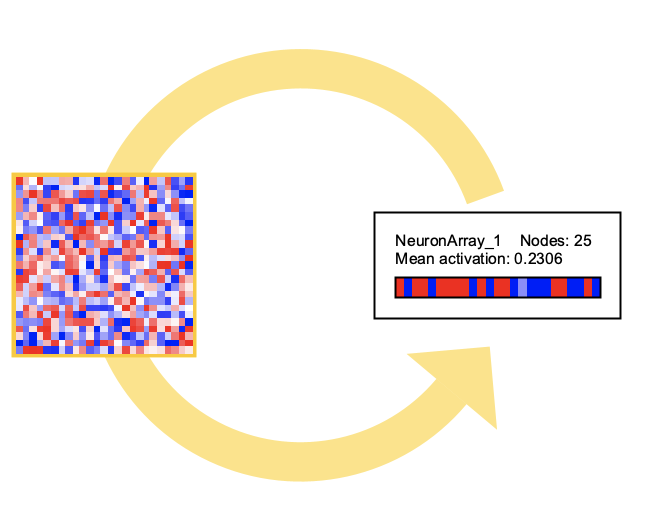
Neuron Arrays
Neuron arrays are collections of neurons backed by Smile matrices, stored as column vectors. They provide efficient computation for large numbers of neurons while maintaining compatibility with Simbrain’s visualization and update systems.
Display Modes
Neuron arrays can be displayed in multiple ways:
- Line mode: Shows neurons as a horizontal or vertical line
- Grid mode: Arranges neurons in a rectangular grid
- Circle mode: Displays individual neurons as circles
- Image mode: Shows activations as a color-coded pixel image
Toggle between these modes using the right-click menu or the GUI tab in the neuron array dialog.
When displaying a non-square neuron array in grid mode, unused pixels in the rectangular grid are rendered as gray.
Array Operations
Neuron arrays support several operations:
- Layer normalization: Normalize activations across the array
- Bias handling: Each neuron can have an individual bias value
- Increment/decrement: Adjust all activations by a fixed amount
- Clear: Reset all activations to zero
- Randomize: Set activations to random values
Activation Sequences
Activation sequences represent sequences of activation vectors, stored as matrices where each row is an activation vector at a particular position in the sequence. Unlike neuron arrays which represent a single vector of activations, activation sequences maintain a temporal or positional dimension.
Activation sequences are not directly accessible from the GUI menu system, but they appear in the network view when used in custom simulations like the Tiny Language Model simulation. When visible, they support standard visualization and editing capabilities.
The key difference from neuron arrays is that activation sequences store multiple activation vectors arranged in sequence (a matrix), enabling parallel processing of sequences at different positions, while neuron arrays represent a single vector of activations suitable for standard feedforward and recurrent layers.
Weight Matrices
Weight matrices represent connections between neuron arrays or neuron groups. They provide efficient matrix-based computation for large-scale networks and support various learning rules and operations.
Matrix Structure
Weight matrices have dimensions of target size × source size. Each entry represents the connection strength from a source neuron to a target neuron. The matrix supports both excitatory and inhibitory connections, with automatic masking to separate these connection types.
Matrix Operations
Weight matrices support numerous operations:
- Diagonalize: Create an identity-like matrix
- Randomize: Fill with random values using the default weight randomizer
- Randomize symmetric: Create symmetric random matrices using the default weight randomizer
- Show weight matrix histogram: Open a dialog to visualize weight distribution and apply weight initialization strategies
- Zero diagonal: Remove self-connections
- Clear: Set all weights to zero
Mathematical Properties
For square matrices, additional operations are available:
- Show eigenvalues: Shows all eigenvalues for this matrix.
- Spectral radius: Set the maximum eigenvalue to control network dynamics. The spectral radius operation scales all weights proportionally so that the maximum eigenvalue equals the specified value. This is done by multiplying the entire weight matrix by the ratio of the desired spectral radius to the current maximum eigenvalue.
Creating Arrays and Matrices
An array can be inserted into a network window through two methods:
Insert > Add Neuron Array- Pressing the keyboard shortcut
Y
Weight matrices can be added by connecting any two neuron arrays or neuron groups. The simplest way to connect two objects is using the 1-2 trick.
Matrix Integration
Simbrain uses efficient matrix operations for array-based computation. This provides:
Benefits
- Fast computation for large networks
- Efficient memory usage
- Modern neural network compatibility
Integration with Other Components
Arrays can connect with other network components:
- Neuron groups can connect to arrays via weight matrices
- Arrays can connect to synapse groups
- Mixed neuron and array networks are supported
PSR Matrix
The Post-Synaptic Response (PSR) matrix is a key innovation in Simbrain that extends traditional matrix-vector multiplication to support spiking neural networks.
Traditional vs PSR Computation
In typical neural networks:
- Output = Weight Matrix × Input Vector
In Simbrain, this is split into two steps:
- PSR Matrix Creation: Each row of the weight matrix is element-wise multiplied by the input vector
- Summation: Row sums of the PSR matrix produce the final output
This factorization allows spike responders to modify individual synaptic responses before summation, enabling more biologically realistic computation.
Spike Response Integration
The PSR matrix can be processed by spike responders before summation, allowing for:
- Non-linear synaptic responses
- Temporal dynamics at individual synapses
- Biologically plausible computation models
Neuron Array Right Click Menu
-
Cut: Cut selected array (Cmd/Ctrl-X)
-
Copy: Copy selected array (Cmd/Ctrl-C)
-
Paste: Paste copied array (Cmd/Ctrl-V)
-
Duplicate: Duplicate selected array (Cmd/Ctrl-D)
-
Edit…: Open properties dialog
-
Delete: Delete selected array (Backspace or Delete)
-
Connect Selected Objects…: Creates weight matrix or synapse group between selected source and target entities
-
Toggle line / grid: Toggle line / grid style (G)
-
Toggle horizontal / vertical layout: Toggle horizontal / vertical layout (L)
-
Toggle circle mode: Toggle activation rendering mode between circle and image (M)
-
Toggle bias visibility: Toggle whether biases are visible (B)
-
Create supervised model: Create supervised model using the current activation as target for immediate training
-
Input Data…: Opens a dialog that can be used to send inputs to this layer
-
Add current pattern to input data: Add the current activation of this layer to the input data table
-
Randomize Selection: Randomize selected elements (r)
-
Randomize Biases: Randomize biases of this neuron array
-
Edit components…: Edit the entries of the activation array in a table
-
Plot Activations (or Plot Activations/Spikes for spiking neurons): Plot neuron activations across available graph types
-
Plot Biases: Plot neuron biases across available graph types
-
Add coupled image world: Couples an image world to this neuron array
-
Record Activations: Start recording activations to a data world
-
Record Biases: Start recording biases to a data world
Weight Matrix Right Click Menu
-
Cut: Cut selected weight matrix (Cmd/Ctrl-X)
-
Copy: Copy selected weight matrix (Cmd/Ctrl-C)
-
Paste: Paste copied weight matrix (Cmd/Ctrl-V)
-
Duplicate: Duplicate selected weight matrix (Cmd/Ctrl-D)
-
Edit…: Open properties dialog
-
Delete: Delete selected weight matrix (Backspace or Delete)
-
Diagonalize: Create an identity-like diagonal matrix
-
Randomize selection: Randomize weights using the default weight randomizer (R)
-
Randomize symmetric: Randomize the matrix symmetrically using the default weight randomizer, ensuring the matrix remains symmetric
-
Show weight matrix histogram…: Open the weight matrix histogram dialog to visualize weight distribution and apply initialization strategies
-
Plot Weight Matrix: Plot weight matrix across available graph types
-
Show eigenvalues…: Show eigenvalues for this matrix (only enabled for square matrices)
-
Set spectral radius…: Rescale matrix so that max eigenvalue is the specified value. The spectral radius operation scales all weights proportionally by multiplying the matrix by the ratio of the desired spectral radius to the current maximum eigenvalue. Values less than 0.9 cause decay; 0.9 churns; greater than 1 explodes
-
Zero diagonal: Effectively removes self-connections (in the recurrent case)
Weight Matrix Histogram Dialog
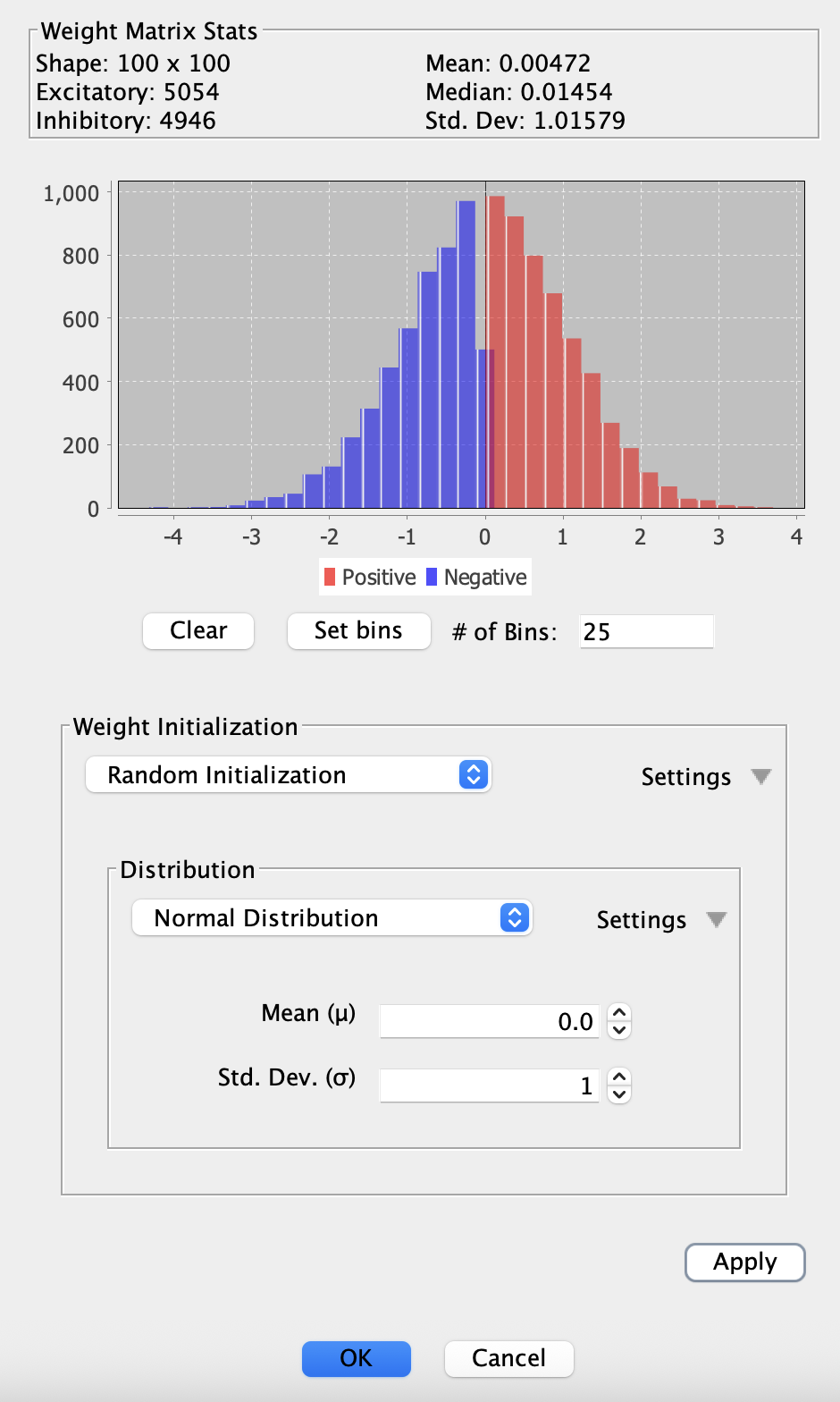
The weight matrix histogram dialog provides visualization and initialization tools for weight matrices. Access it by right-clicking a weight matrix and selecting Show weight matrix histogram....
Statistics Panel
The top section displays key statistics about the weight matrix:
- Shape: Dimensions of the matrix (rows × columns)
- Mean: Average weight value across all connections
- Excitatory: Count of positive weights
- Median: Middle value when all weights are sorted
- Inhibitory: Count of negative weights
- Std. Dev: Standard deviation of weight values
Histogram Display
The histogram shows the distribution of weight values from negative to positive. Positive weights appear in red, while negative weights appear in blue, making it easy to visualize the balance between excitatory and inhibitory connections.
Weight Initialization Strategies
The bottom section allows you to apply different weight initialization strategies commonly used in machine learning. Select a strategy from the dropdown and click Apply to reinitialize the matrix:
-
Random Initialization: Apply any probability distribution to the weights. This is the most flexible option, allowing you to use Uniform, Normal, Poisson, or any other available distribution
-
Xavier: Initializes weights using Xavier/Glorot initialization, which helps maintain activation variance across layers. Works well for sigmoid and tanh activations. Choose between Uniform or Normal distribution variants
-
He: Optimized for ReLU activation functions. Scales weights based on the number of input connections. Choose between Uniform or Normal distribution variants
-
LeCun: Classic initialization method that scales weights based on the number of inputs. Designed for SELU activation functions. Choose between Uniform or Normal distribution variants
After applying a strategy, the histogram and statistics update immediately to reflect the new weight distribution. This allows you to experiment with different initialization approaches and see their effects in real-time.
For more information on when to use each strategy, see Training Parameters.
Randomization and Initialization
Weight matrices can be randomized in several ways:
Quick Randomization
The Randomize selection and Randomize symmetric menu items use the default weight randomizer configured in network preferences. This randomizer applies to all weights regardless of polarity and can change the sign of weights.
To use these options:
- Right-click the weight matrix
- Select
Randomize selection(or press R) for standard randomization - Select
Randomize symmetricfor symmetric matrices (useful for recurrent networks)
Both options use the same randomizer configured in File > Network Preferences > Randomizers > Weight randomizer.
Advanced Initialization
For more control over weight initialization, use the weight matrix histogram dialog. This dialog provides:
- Visual feedback through the histogram
- Access to specialized initialization strategies (Xavier, He, LeCun)
- Ability to configure custom probability distributions
- Real-time statistics updates
The histogram dialog is particularly useful when setting up networks for machine learning tasks, where proper weight initialization can significantly impact training performance.
Differences with Synapse Groups
While weight matrices and synapse groups both represent connections, they have different capabilities:
Weight Matrices
- Fast computation for large networks
- Best for fully-connected layers
- Support matrix operations (eigenvalues, spectral radius)
- Learning rules work on entire matrix
Synapse Groups
- Individual synapse properties
- Support connection delays
- More learning rule options
- Can connect any network components
- Support for sparse connectivity
Weight Matrix Dialog Options
Properties Tab
- Label: Optional string description for identification
- Update Priority: Determines update order in network (lower numbers update first)
- Increment amount: Amount to increment all weights when using keyboard shortcuts
- Clamped: If checked, local learning rules won’t be applied (weights can still be manually updated)
- Learning rule: Specifies how weights change during training
- Spike responder: Processes PSR matrix entries, only used if source connector’s rule is spiking (see Spike Responders)
Weight Matrix Tab
The weight matrix viewer allows you to view and edit weight values in a tabular format, where cells represent connections between source neurons (rows) and target neurons (columns). Values can be edited directly, randomized, or loaded/saved from .csv files.
Data Tab
- Learning rule data: State variables for the learning rule (if any)
- Spike responder data: State variables for the spike responder (if any)
- PSR matrix: View post-synaptic response values
Neuron Array Dialog Options
Main Tab
- Activations: Current activation values for all neurons
- Label array: Individual labels for each neuron in the array
- Bias Array: Individual bias values for each neuron
- Use Layer Norm: If checked, applies layer normalization after each update
- Label: Optional string description for identification
- Update Priority: Determines update order in network (lower numbers update first)
- Clamped: Prevents activation updates (see clamping)
- Increment: Amount to increment all activations using keyboard shortcuts
GUI Tab
- Grid Mode: Show neurons in rectangular grid vs. single line
- Vertical Layout: Orient array vertically instead of horizontally
- Circle Mode: Display individual neuron circles instead of color-coded image
- Show Bias: Show bias values as small indicators
Update Rule Tab
- Update Rule: Neuron update function (Linear, Sigmoidal, etc.)
- State variables: Rule-specific state variables (depends on selected update rule)
Update rule parameters vary by rule type. For more information on specific update rules, see neuron update rules.