Neuron Groups and Collections
Neuron groups and collections are two ways to organize neurons in Simbrain. Both appear as sets of neurons surrounded by a gray outline with an interaction box, but they serve different purposes.
Neuron groups are structural containers where neurons are children of the group. They enforce consistency, requiring all neurons to share the same update rule type. Groups support specialized types like SOM, Competitive, and WTA, and are commonly used for layer organization in feed-forward networks and structured architectures. Create a neuron group using the Insert menu or by checking the group option when adding neurons. Neuron groups have a yellow interaction box and are also constituents of subnetworks. Deleting a group removes all its neurons.
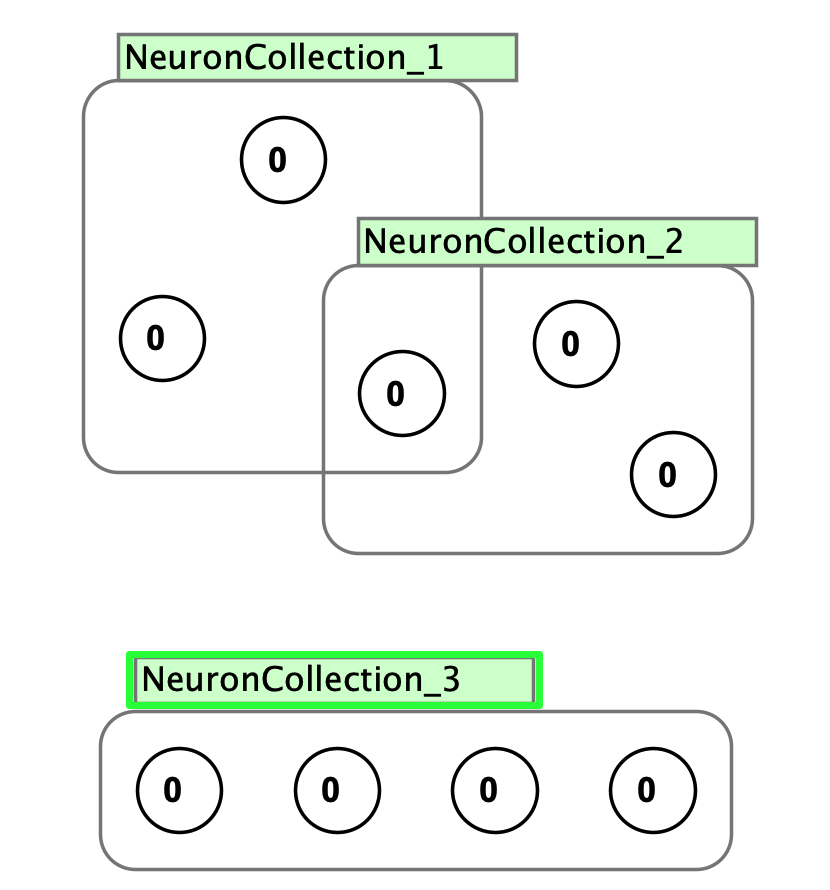
Neuron collections are lightweight groupings primarily for labeling and visual organization. They allow flexible membership where neurons can belong to multiple collections simultaneously and can have different update rules. Create a collection by selecting neurons and pressing Cmd/Ctrl+G. Collections have a green interaction box. Deleting a collection leaves the neurons intact.
Both groups and collections can be connected using weight matrices or synapse groups. Use the 1-2 trick for matrix-based connections or the 1-3 trick for individual synapse management.
Creation/Edit Dialog
When creating or editing a neuron group, several common properties are available:
- Group ID: Unique identifier for the group
- Label: Display name for the group
- Population: Number of neurons in the group (read-only after creation)
- Neuron Type: The update rule governing neurons in the group
- Parent: The parent network or subnetwork, if any
Additional tabs provide access to:
- Neurons: Configure individual neuron properties
- Layout: Arrange neurons spatially
- Input Data: Set up input patterns for training or testing
Group-specific parameters vary by type. See individual group type pages for details on specialized parameters like learning rates, temperature, or winner values.
Neurons
See the neuron page.
Layout
See the layouts page.
Input Data
Neuron groups and other layers can accept input data through input tables. Right-click on a layer and choose Input data to open an input table interface.
Right Click Menu
The context menu for neuron groups and collections provides access to common operations. The menus are identical except for minor wording differences (neuron groups say “Select Internal Neurons” and “Remove Neuron Group” while collections say “Select Neurons” and “Remove Neuron Collection”).
Basic Operations:
- Edit…: Opens the group properties dialog (same as double-clicking the interaction box)
- Rename Neuron Collection…: Change the group’s display name
- Remove Neuron Group / Remove Neuron Collection: Delete the group/collection and all its neurons (for groups only)
- Apply layout…: Rearrange neurons using different layout algorithms (keyboard shortcut:
Cmd/Ctrl+L)
Selection:
- Select Internal Neurons / Select Neurons: Select all neurons in the group or collection (keyboard shortcut:
S) - Edit Neurons…: Opens an editor dialog for all neurons. When multiple neurons are selected, the dialog title will show the count (e.g., “Edit 4 Neurons”)
- Select Incoming Synapses: Select synapses targeting this group
- Select Outgoing Synapses: Select synapses originating from this group
Clamping:
- Clamp Neurons: Clamp all neurons (see Clamping)
- Unclamp Neurons: Unclamp all neurons
Connecting Groups:
- Connect selected objects with weight matrix: Connect to other groups using weight matrices
- Connect selected neuron groups with synapse group: Connect using synapse groups
- Set Group as Source: Mark this group as source for connection operations
- Clear Source Neuron Groups: Clear source selection
Training and Testing:
- Create supervised model: Set up supervised learning using current activations as targets (keyboard shortcut:
Cmd/Ctrl+M) - Input data…: Open dialog to configure input patterns
- Add current pattern to input data…: Add current activations to the input data table
Visualization and Analysis:
- Plot: Create a plot of the group’s activations over time
- Add coupled image world: Create an image world coupled to this group
- Record activations: Record activation values to a CSV file
Couplings:
- Send Vector Coupling To: Send activations as a vector to other Simbrain components
- Receive Vector Coupling from: Receive vectors from other components to set activations
Some specialized group types (like SOM) have additional custom menu items such as Reset SOM Network, Recall SOM Memory, and Randomize SOM Weights.
See the couplings page for more information on connecting neuron groups to other Simbrain components.